T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy that frequently occurs in children, adolescents, and young adults. Approximately 10-20% of T-ALL patients will experience relapse months or years following remission and will often become refractory to further treatments. The survival of relapsed/refractory patients is very poor, with an overall survival rate of less than a 25% overall survival rate. Relapsed patients often have enriched pools of leukemia initiating cells (LICs) with enhanced pro-survival and self-renewal capacity, suggesting a potential vulnerable population for effective targeted therapies with less toxicity.
An emerging research topic in LIC biology is the identification of RNA modifying enzymes that are important for LIC self-renewal and survival. ADAR1 enzymes catalyze the transition of adenosine (A) to inosine (I) in precursor double-stranded RNA (dsRNA). Epitranscriptomic A-to-I RNA editing events are widespread in the cancer transcriptome and are critical for the transition from pre-leukemic cells to fully functional LICs. Compared to myeloid leukemia, the role of ADAR1 in lymphoid progenitor maintenance and malignant transformation is not well understood .
A-to-I RNA editing has a wide range of effects on RNA biology including gene expression, splicing, RNA degradation and translation, and miRNA biogenesis and/or 3' UTR targeting. The best documented functional roles of ADAR1 are suppression of the interferon (IFN) response and RNA editing of self-dsRNA to prevent abnormal activation of cytosolic self-dsRNA sensing. Concurrent deletion of the cytosolic dsRNA sensors melanoma differentiation-associated protein 5 (MDA5) and protein kinase R (PKR) is able to completely rescue embryo death and reverse the IFN signatures. Whether editing of immunogenic dsRNA and suppression of aberrant dsRNA sensing pathway could enhance LIC self-renewal capacities is an important question that has not been extensively addressed.
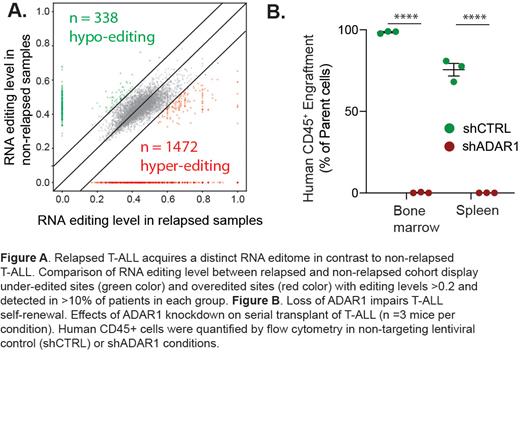
In this study, we applied bioinformatic analysis on a large cohort of T-ALL samples to examine the function of ADAR1 in the context of T-ALL LIC maintenance. We found that ADAR1 is highly expressed in ~70% of all T-ALL patients and particularly within the LIC compartment. A thorough comparison of the A-to-I RNA editing landscape between non-relapsed and relapsed T-ALL patient cohorts revealed hyper-editing is associated with both increased risk of relapse and leukemia-associated mortality. A total of 338 under-edited and 1,472 over-edited sites were found in relapsed patients compared to non-relapsed samples. However, there was very little difference in ADAR1 expression and the overall A-to-I RNA editing levels, among various molecular subtypes of T-ALL, suggesting malignant A-to-I RNA editing is a common attribute of relapsed T-ALL regardless of the genetic mutation status.
We also performed functional study in a three-dimensional human thymic organoid system and a T-ALL patient-derived xenograft (PDX) model. Depletion of ADAR1 showed striking effects in LIC survival and self-renewal with 50-90% reduction in leukemia growth. Mechanistically, we revealed complex dsRNA regulatory mechanisms of ADAR1 by directing hyper-editing of immunogenic dsRNA and retains unedited nuclear dsRNA to avoid detection by the innate immune sensor MDA5. Interesting, the dependency on ADAR1-MDA5 axis various among patients depending on the cell intrinsic level of MDA5. Collectively, our results show that ADAR1 functions as a self-renewal factor that limits the sensing of endogenous dsRNA. Thus, targeting ADAR1 presents a safe and effective therapeutic strategy for eliminating T-ALL LICs.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal